Technology
What are Aptamers? Aptamers are DNA/RNA molecules that have affinities for their targets similar to antibodies. Aptamers have shown great potential to replace antibodies in biosensors, point-of-care diagnostics, therapeutics, and all of the areas currently dominated by antibodies. Once aptamers have been identified, they can be produced for less than 10% of the cost of antibodies. The commercial value of antibodies exceeds $40 billion per year. AptaMatrix continues to explore innovative approaches towards aptamer development.
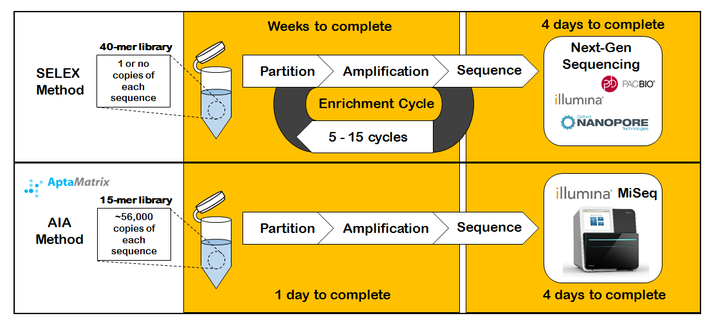
How are aptamers generated? Traditionally DNA aptamers have been generated using the Systemic Evolution of Ligands by Exponential Enrichment method (called SELEX). This cyclic evolution approach uses a random DNA library (40-70 nucleotides) screened against a protein target of interest. Commercial success of aptamers has been limited by the cumbersome SELEX method for aptamer discovery, as only a few hundred viable aptamers have been discovered in the last 20 years, compared to thousands of antibodies over the same time period.
What makes AptaMatrix's approach different? In 2011, AptaMatrix focused their efforts on developing innovative, non-SELEX approaches to aptamer discovery. With the arrival of next-gen sequencing instrumentation in 2010 (i.e. 454 and Illumina GAiix), AptaMatrix pioneered the use of next-gen sequencing technologies towards accelerating aptamer discovery. This work culminated into was is now termed our Acyclic Identification of Aptamers™ (AIA™) method for aptamer discovery (formerly called High-Throughput Screening of Aptamers (HTSA), PLOS ONE 2011). The AIA method leverages (1) short, over represented DNA aptamer libraries (15-25 nucleotide), (2) innovative screening/partitioning conditions, (3) efficient adapter ligation methods and (4) next-gen Illumina sequencing instrumentation to dramatically reduce the cost and time to discover new aptamers, from months to days. The AIA approach aims to streamlining the discovery of aptamers against protein targets of commercial interest.
What are Aptamers? Aptamers are DNA/RNA molecules that have affinities for their targets similar to antibodies. Aptamers have shown great potential to replace antibodies in biosensors, point-of-care diagnostics, therapeutics, and all of the areas currently dominated by antibodies. Once aptamers have been identified, they can be produced for less than 10% of the cost of antibodies. The commercial value of antibodies exceeds $40 billion per year. AptaMatrix continues to explore innovative approaches towards aptamer development.
How are aptamers generated? Traditionally DNA aptamers have been generated using the Systemic Evolution of Ligands by Exponential Enrichment method (called SELEX). This cyclic evolution approach uses a random DNA library (40-70 nucleotides) screened against a protein target of interest. Commercial success of aptamers has been limited by the cumbersome SELEX method for aptamer discovery, as only a few hundred viable aptamers have been discovered in the last 20 years, compared to thousands of antibodies over the same time period.
What makes AptaMatrix's approach different? In 2011, AptaMatrix focused their efforts on developing innovative, non-SELEX approaches to aptamer discovery. With the arrival of next-gen sequencing instrumentation in 2010 (i.e. 454 and Illumina GAiix), AptaMatrix pioneered the use of next-gen sequencing technologies towards accelerating aptamer discovery. This work culminated into was is now termed our Acyclic Identification of Aptamers™ (AIA™) method for aptamer discovery (formerly called High-Throughput Screening of Aptamers (HTSA), PLOS ONE 2011). The AIA method leverages (1) short, over represented DNA aptamer libraries (15-25 nucleotide), (2) innovative screening/partitioning conditions, (3) efficient adapter ligation methods and (4) next-gen Illumina sequencing instrumentation to dramatically reduce the cost and time to discover new aptamers, from months to days. The AIA approach aims to streamlining the discovery of aptamers against protein targets of commercial interest.
What is AptaMatrix doing with aptamers? Once an aptamer is validated (checked for specificity and affinity to its protein target), the sequence is patented and useful for a variety of applications that parallel that of antibodies (biosensing, affinity purification, target capture, therapeutics, etc.). AptaMatrix has extensive experience engineering short aptamers into DNA biosensors for near real-time protein detection, called AlloSwitch™ sensors. The AlloSwitch™ technology has demonstrated the ability to transduce molecular recognition of biological targets into a near real-time (seconds) optical signal for identifying environmental and terrorist released contaminants in liquid systems and for high-throughput drug discovery applications.
AptaMatrix has recently made significant improvements to the existing AIA method to enabling massively parallel screening of heterogeneous protein mixtures. This improved method, called Multiplexed AIA™ (mAIA™), has the potential to screen 1000's of proteins against a collection of aptamer libraries (15-25 nucleotides) in a single screen (i.e. single tube). Once optimized, a single mAIA screen has the potential to identify more DNA aptamers in a single screen, than what has been published in the last 20 years. The mAIA method will allow AptaMatrix to rapidly advance aptamer-based applications in disease detection.
AptaMatrix has recently made significant improvements to the existing AIA method to enabling massively parallel screening of heterogeneous protein mixtures. This improved method, called Multiplexed AIA™ (mAIA™), has the potential to screen 1000's of proteins against a collection of aptamer libraries (15-25 nucleotides) in a single screen (i.e. single tube). Once optimized, a single mAIA screen has the potential to identify more DNA aptamers in a single screen, than what has been published in the last 20 years. The mAIA method will allow AptaMatrix to rapidly advance aptamer-based applications in disease detection.